Crop Wild Relatives Mining
My research focuses on harnessing genetic diversity from crop wild relatives (CWR) to enhance traits for crop improvement, with an emphasis on wheat and mint. In wheat, I have explored vernalization genes to identify natural genetic variations in ancient wheats and wild relatives, enabling adaptations such as spring growth habits. A key finding, published in 2015 in Proceedings of the National Academy of Sciences, traced the origin of spring growth in ancient South Asian wheats to the VERNALIZATION 4 (VRN4) gene. By leveraging genetic diversity from wild and ancient accessions, I mapped alleles that reduce vernalization requirements, focusing on epistatic interactions and polymorphisms in loci like VRN-D4 and VRN-A1. This work supported the development of winter cereals adapted to diverse climates using germplasm collections.
In mint, an aromatic crop, I conducted surveys of the U.S. National Plant Germplasm System (NPGS) collection from 2018 to 2021, identifying novel peppermint-type oil sources from wild relatives such as Mentha longifolia. This research validated breeding strategies to boost oil yields by incorporating induced and natural genetic variation from wild accessions. The result was the development of varieties like 'Velvet' peppermint, which offers higher yields and is now commercially produced. My approach integrates germplasm characterization for traits like disease resistance and yield, utilizing field trials and mutant populations to develop pre-breeding materials adopted across thousands of acres for commercial use.
Genomic Tools Development in Orphan Crops
I have played a key role in developing genomics tools for orphan crops—under-researched species like mint and chickpeas—facilitating accelerated breeding through molecular markers, genome assemblies, and genotyping platforms. At UC Davis, in collaboration with Mars Inc., I spearheaded genomics for perennial crops like mint, an orphan crop with limited prior tools. Notable achievements include the 2024 chromosome-scale assembly of apple mint (Mentha suaveolens), which integrated sequencing methods for improved de novo assembly, and the 2021 efficient linkage map for M. suaveolens using sequence capture to identify haplotypes. These tools enabled gene discovery for flavor, yield, and disease resistance, leading to diploid mint varieties producing spearmint-type oil with high yield potential.
My work also involved deploying SNP genotyping platforms and molecular markers in polyploid crops, optimizing assays for genome duplication and pre-breeding. At NuCicer Inc., as Plant Breeding Director, I advanced genomics for chickpeas (often classified as an orphan crop in breeding contexts), developing roadmaps for varietal selection, predictive breeding, and automated phenotyping. This included managing projects with external partners to enhance genetic diversity using tools like high-throughput sequencing and statistical analysis in R and SAS. My technical skills in DNA technologies, such as designing markers for diploid and polyploid species, have been instrumental in characterizing natural variation and producing pre-breeding materials.
Environmental fitness and product placement
My career emphasizes enhancing environmental fitness—crop adaptation to abiotic and biotic stresses—and strategic product placement, ensuring varieties perform optimally in target environments through field-based evaluation and interdisciplinary breeding. In wheat, my research on phytochrome (phyB and phyC) mutations and vernalization pathways addressed environmental cues like photoperiod and temperature, revealing shared functions in flowering and shade-avoidance. RNA-seq studies under varying day lengths identified gene networks for reproductive fitness, enabling wheat lines with no functional VRN2 genes for better adaptation to short/long days and climate variability. This work supports product placement by tailoring varieties to specific latitudes and seasons, maximizing grain production. In mint and chickpeas, I conducted field trials at sites like UCANR's Extension Centers (IREC) to test environmental performance under California conditions, replicating farming stresses like pests and variable climates. As Plant Breeding Lead at NuCicer, I developed strategies for product advancement, focusing on fitness traits like yield, disease resistance, and resource efficiency, while collaborating across analytics, bioinformatics, and R&D for market-aligned placement.
Techniques
Throughout the years I spend time implementing a different variety of techniques on plant research and variety development:- Colchicine whole chromosome doubling
- Tilling using EMS (Ethyl Methyl Sulfonate) in seed crops
- Gamma radiation to increase genetic diversity in clonal crops
- Speed Breeding
- Near-Infrared (NIR) Spectroscopy for quality traits
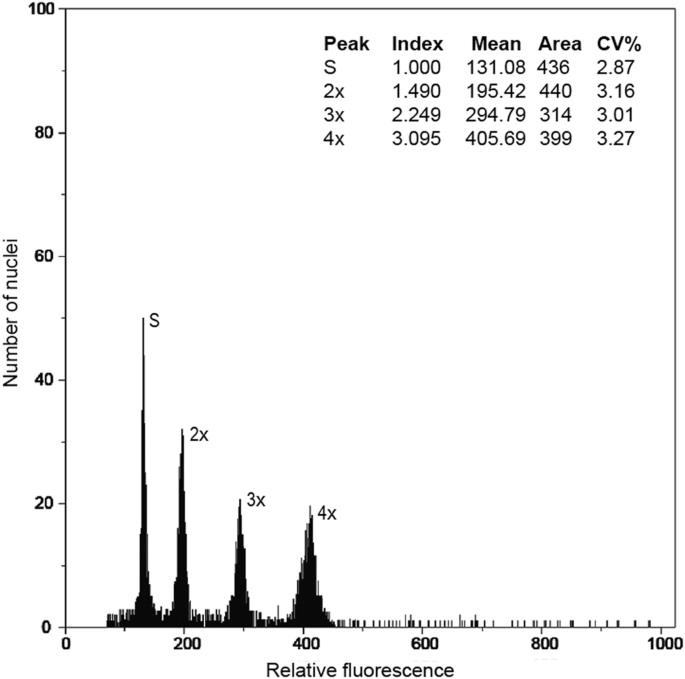
- Flow Cytometry Analysis for genome size estimations
- Hyperspectral Imaging (HSI) for phenotyping
- UAV phenotyping (for rapid assessment of aerial traits)
- Illumina High throuput Sequencing for variant discovery
- Illumina SNP arrays for trait discovery, mapping, and genomic prediction
- PacBio HiFi
- Transcriptomic analysis using Illumina Sequencing
- Transcripts database construction using PacBio IsoSeq
- KASP genotyping for rapid SNP detection
- Protoplast extraction
- Plasmid delivery for transient gene expression
- Gas Chromatography (GC) for trait quality
- Ultra High-Performance Liquid Chromatography for rapid phenotyping
- Lab-based methods for assay validation (such as Soxhlet for oil extraction)